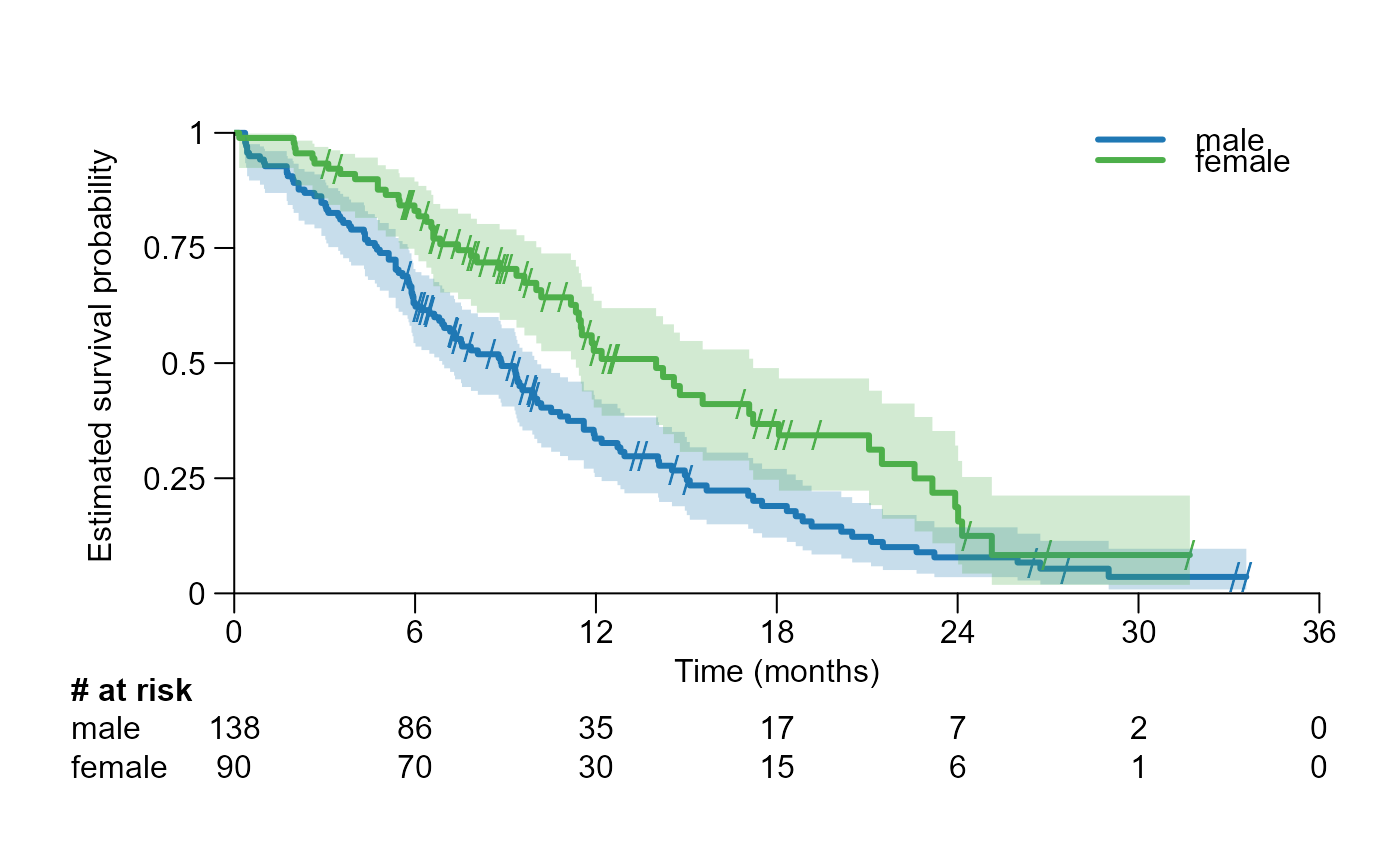
Provide an open-source, user-friendly tool designed to enhance the creation
and customization of Kaplan-Meier plots and incorporating various statistics
and layout customization options using surv.plot(fit, ...).
Arguments
- fit
An object of class survival::survfit containing survival data.
- ties
String specifying the method for tie handling. If there are no tied death times all the methods are equivalent. Options: "efron", "breslow", "exact". Default: "efron"
- reference.arm
A character string specifying the reference arm for comparison.
- time.unit
A character string specifying the time unit which was used to create the
fitobject. Note:time.unitwill not convert the time of thefitobject.Option include:
"day","week","month","year".- y.unit
A character string specifying the unit of the y-axis.
Option include:
"probability","percent".- censoring.mark
A logical parameter indicating whether censoring events should be marked on the survival curves. Default is
TRUE.- censoring.cex
A numeric value specifying the size of the marks for censored patients. Default is
1.3.- conf.int
A numeric value controlling the confidence interval on survival curves. Default is
0.95, corresponding to a 95% confidence interval. Values between0and1represent the desired confidence interval. If set to0, no confidence intervals are displayed.- conf.band
A logical parameter indicating whether to display the confidence band on the survival curves. Default is
TRUE.- conf.band.col
A colour which is used for the confidence band. Can accept a single colour value or a vector of colours.
- conf.band.transparent
A numeric value between
0and1controlling the transparency of the confidence band. Default is0.25.- conf.line.lty
A strings specifying the line type of the confidence lines.
Options include:
"blank","solid","dashed","dotted","dotdash","longdash","twodash". Default is"blank".- conf.line.lwd
A numeric value specifying the width of the confidence lines.
- conf.type
Transformation type for the confidence interval.
Options include:
"log","log-log","plain","logit","arcsin". Default islog-log.- grid
A logical parameter specifying whether to draw a grid. Default is
FALSE.- col
A colour which is used for the survival curves. Can accept a single colour value or a vector of colours.
- main
Title of the plot.
- sub
Subtitle of the plot. Note: A subtitle is only displayed if no risk table is shown.
- letter
A letter to be displayed on the top to the left of the figure (for example if several plots are displayed in one figure).
- letter.cex
The size of
letterwhich is displayed on the top to the left.- letter.pos.x
The X-axis position of
letter(corresponds toadjinmtext). Default is -0.35- letter.pos.y
The Y-axis position of
letter(corresponds tolineinmtext). Default is 1.- xlab
X-axis label.
- ylab
Y-axis label.
- xticks
A numeric vector specifying the ticks of the x-axis.
Can be specified as
seq(from = , to = , by = ).from: starting valueto: end valueby: number; increment of the sequence
- yticks
A numeric vector specifying the ticks of the y-axis.
Can be specified as
seq(from = , to = , by = ).from: starting valueto: end valueby: number; increment of the sequence
Note: It should always be specified as probability.
- xlab.pos
Defines the margin line where the X-axis label (xlab) is displayed, starting at 0 and counting outwards. Default is 1.5.
- ylab.pos
Defines the margin line the Y-axis label (ylab) is displayed, starting at 0 counting outwards. Default is 3.
- xlab.cex
A numeric value specifying the size of the X-axis label.
- ylab.cex
A numeric value specifying the size of the Y-axis label.
- cex
A numeric value specifying the size of all all text elements (labels, annotations, etc.).
- axis.cex
A numeric value specifying the size of the axis elements.
- bty
Determines the style of the box drawn around the plot.
Options include:
"n","o","c","u". Default is"n".- lty
A string specifying the line type of of the curve(s).
Options include:
"blank","solid","dashed","dotted","dotdash","longdash","twodash".- lwd
A numeric value specifying the width of the line.
- legend
A logical parameter specifying whether to display legend. By default, the legend is displayed if there is more than one arm.
- legend.position
Position of the legend.
Options include: "c(x,y)"
,"bottomright","bottom","bottomleft","left"`,- legend.name
A vector of character string specifying of the name(s) of the arm(s).
- legend.cex
A numeric value specifying the size of the legend text.
- legend.text.font
Font style of the legend text. Possible values:
1normal2bold3italic4bold and italic
- legend.title
Title of the legend.
- legend.title.cex
A numeric value specifying the size of the legend title.
- segment.type
A numeric value specifying the layout of the segment. Possible values:
1full width2half width3vertical and horizontal segment (default)
- segment.timepoint
A single value or a vector of fixed time points to be drawn as segment(s).
- segment.quantile
A single value or a vector of fixed quantile to be drawn as segment(s). Example: 0.5 corresponds to median.
- segment.main
Title of the segment text.
- segment.confint
A logical parameter specifying whether to display the confidence interval for the segment. Default: TRUE
- segment.annotation
Position of the segment annotation. If several quantiles or time points are chosen then the position can only be set to
"left","right","top"or"bottom".Options include:
c(x,y),"bottom","bottomleft","left","right","top","none".- segment.annotation.offset
The offset used in function
text()for adding the annotation if several time points or quantiles are added.- segment.annotation.two.lines
A logical parameter to force that the annotation is displayed on two lines. This parameter only has an effect if there is only one arm. Default: FALSE
- segment.annotation.col
A colour which is used for the segment annotation. Can accept a single colour value or a vector of colours.
- segment.annotation.space
Spacing between the text in units of y-coordinates.
- segment.col
A colour which is used for the segment. Can accept a single colour value.
- segment.lty
A strings specifying the line type of the segment(s).
Options include:
"blank","solid","dashed","dotted","dotdash","longdash","twodash".- segment.lwd
A numeric value specifying the width of the segment line.
- segment.cex
A numeric value specifying the size of the segment text size.
- segment.font
A numeric value specifying the font face. Possible values:
1plain2bold3italic4bold-italic
- segment.main.font
A numeric value specifying the font face for the segment text. Possible values:
1plain2bold3italic4bold-italic
- stat.fit
An object of class survival::survfit containing survival data. Used for calculation of statistics, allowing to add stratification factors.
Note: If not specified the
fitobject will be used for thestat.- stat
Statistics displayed in the plot.
Options:
"logrank"gives the p value of the conducted logrank test usingsurvdiff{survival}. To tests if there is a difference between two or more survival curves."coxph"gives the hazard ratio (HR) and its CI (default: 95% CI)of the conducted Cox proportional hazards regression usingcoxph{survival}. Note: This option only works if there are two arms."coxph_logrank"combines the hazard ratio (HR), its CI (default: 95% CI) and the logrank test. Note: This option only works if there are two arms.'none'no statistic is displayed (default).
Note: Confidence interval can be adjusted with the argument
stat.conf.int.- stat.position
Position where the
statshould be displayed.Options include:
c(x,y),"bottomleft","left","right","top","topright","bottomright","none".- stat.conf.int
A numeric value controlling the confidence interval on the
stat(hazard ratio). Default is0.95, corresponds to a 95% confidence interval. Values between0and1represent the desired confidence interval.- stat.col
A colour which is used for the statistics text. Can accept a single colour value or a vector of colours.
- stat.cex
A numeric value specifying the size of the `statistics text size.
- stat.font
The font face of the statistics Possible values:
1plain2bold3italic4bold-italic
- risktable
A logical parameter indicating whether to draw risk table. Default is
TRUE.- risktable.censoring
A logical parameter indicating whether to display number of censored patients. Default is
FALSE.- risktable.pos
Defines on which margin line of the xlab is displayed, starting at 0 counting outwards. Default is at line
3.- risktable.name
Names of the arms for the risk table.
- risktable.cex
A numeric value specifying the size of the risk table text size.
- risktable.col
A coulour which is used for the risk table. Can accept a single colour value or a vector of colours. Default is
black.Note: If
risktable.col = TRUEthen the colours of the curves are used.- risktable.title
Title of risk table.
- risktable.title.font
Font style of the risk table. Possible values:
1normal2bold3italic4bold and italic
- risktable.title.col
A colour which is used for the risk table title. Can accept a single colour value.
- risktable.title.position
A numeric value specifying the position of the title on the x-axis.
- risktable.title.cex
A numeric value specifying the size of the risk table title size.
- risktable.name.cex
A numeric value specifying the size of the risk table legend name size.
- risktable.name.font
Font style of the risk table legend name(s). Possible values:
1normal2bold3italic4bold and italic
- risktable.name.col
A colour which is used for the risk table name. Can accept a single colour value.
- risktable.name.position
A numeric value specifying the position of the legend name(s) on the x-axis.
- margin.bottom
Specifies the bottom margin of the plotting area in line units. Default is
5.- margin.left
Specifies the left margin of the plotting area in line units. Default is
6(with risktable) or4(without risktable).- margin.top
Specifies the top margin of the plotting area in line units. Default is
3.- margin.right
Specifies the right margin of the plotting area in line units. Default is
2.- theme
Built-in layout options. Options include: ("none", "SAKK", "Lancet", "JCO", "WCLC", "ESMO")
Details
The survSAKK R package provides the surv.plot() function, facilitating
Kaplan-Meier survival analysis. Designed with user-friendliness and efficiency
in mind. Offering robust tool for analysing survival data. It utilises the
functionalities of survival::survfit().
For a comprehensive manual visit: https://sakk-statistics.github.io/survSAKK/articles/surv.plot.html
References
Therneau T (2024). A Package for Survival Analysis in R. R package version 3.5-8, https://CRAN.R-project.org/package=survival.
Terry M. Therneau, Patricia M. Grambsch (2000). Modeling Survival Data: Extending the Cox Model. Springer, New York. ISBN 0-387-98784-3.
See also
survival::survfit()which this function wraps.